M. Shahidul Islam, PhD

Associate Professor, Department of Chemistry
Email: sislam [at] desu.edu | Office: 302.857.7181
Website
EDUCATION
- Postdoctoral Fellow: University of Chicago, 2016
- Postdoctoral Fellow: University of Waterloo
- Ph.D., Chemistry, Memorial University, 2008
- MBA, University of Illinois at Chicago, 2022
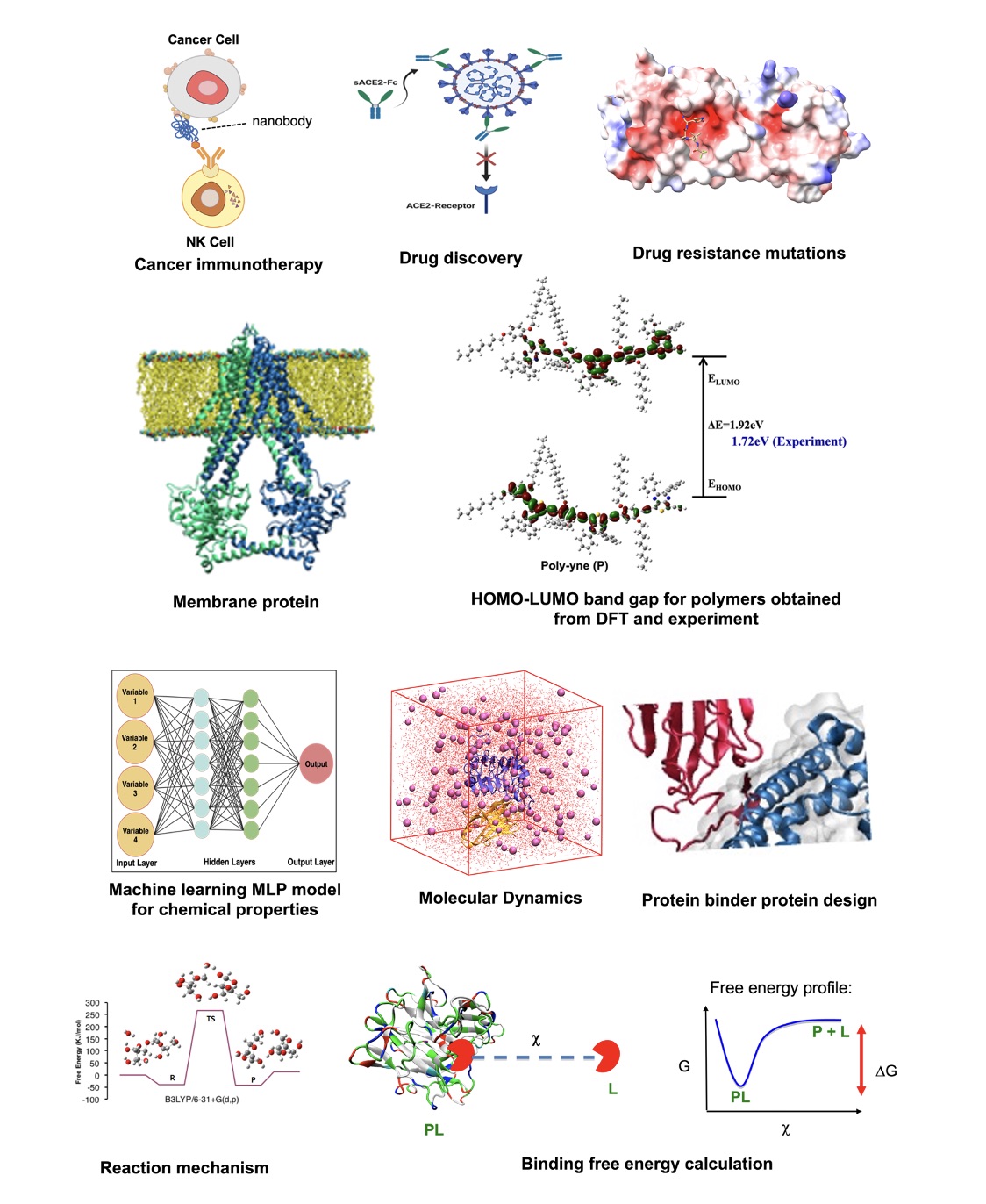
RESEARCH INTERESTS
- Computationally aided drug discovery
- Infectious disease such as COVID-19
- Immunotherapeutics for Cancer
- Genetic diseases such as PKU
- De novo protein and peptide design by AI, deep learning tools
- Understanding structure, dynamics, and function of membrane proteins
- Development of affordable polymers for opto-electronic (O-E) applications, such as solar cells, using DFT calculations and AI/machine learning approach
SELECTED PUBLICATIONS
Dr. Islam has published over 50 peer-reviewed journal papers. Some selected publications are as follows:
- Lopez UM, Hasan MM, Havranek B, Islam SM. SARS-CoV-2 Resistance to Small Molecule Inhibitors. Curr. Clin. Microbiol. Rep. Published online June 24, 2024:1-13. https://doi.org/10.1007/s40588-024-00229-6
- Havranek B, Demissie R, Lee H, Lan S, Zhang H, Sarafianos S, Ayitou AJ, Islam SM. Discovery of Nirmatrelvir Resistance Mutations in SARS-CoV-2 3CLpro: A Computational-Experimental Approach. J Chem Inf Model. 2023 Nov 27;63(22):7180-7188. https://doi.org/10.1021/acs.biochem.3c00561
- Havranek, B.; Lindsey, G. W.; Higuchi, Y.; Itoh, Y.; Suzuki, T.; Okamoto, T.; Hoshino, A.; Procko, E.; Islam, S. M., “A Computationally Designed ACE2 Decoy Has Broad Efficacy against SARS-CoV-2 Omicron Variants and Related Viruses in Vitro and in Vivo”. Commun. Biol. 2023, 6 (1), 513. https://doi.org/10.1038/s42003-023-04860-9
- Havranek, B.; Demissie, R.; Lee, H.; Lan, S.; Zhang, H.; Sarafianos, S.; Ayitou, A. J.-L.; Islam, S. M., Discovery of Nirmatrelvir Resistance Mutations in SARS-CoV-2 3CLpro: A Computational-Experimental Approach. J. Chem. Inf. Model. 2023, 63 (22), 7180–7188. https://doi.org/10.1021/acs.jcim.3c01269
- Abousamra, W. H.; Thomas, D.; Yang, D.; Islam, S. M.; Winstead, C.; Kim, Y.-G. Synthesis and Characterization of the Donor-Acceptor Conjugated Polymer PBDB-T Implementing Group IV Element Germanium. Polymers. 2023, 15 (11), 2429. https://doi.org/10.3390/polym15112429
- Almanasra, A. Havranek, B.; Islam, S.M. “In silico screening and molecular dynamics simulation reveal single point mutations that destabilize β-hexosaminidase A (HexA) causing Tay-Sachs disease”, Proteins, 2021, 89(11), 1587-160. https://doi.org/10.1002/prot.26180
- Havranek, B.; Chen, K., Wu, A.; Procko, E.; Islam, S.M. “Computationally designed ACE2 decoy receptor binds SARS-CoV-2 spike (S) protein with tight nanomolar affinity” J. Chem. Inf. Model., 2021, 61(9), 4656-4669. https://doi.org/10.1021/acs.jcim.1c00783
- Al-Busaidi, I.J.; Haque, Balushi, R.; A.; Rather, J. A; Munam, A.; Islam, S.M., Wong, W-Y.; Skelton, J. M.; Khan, M.S. “Synthesis, characterization, and optoelectronic properties of phenothiazine-based organic co-poly-ynes” New J. Chem., 2021, 45, 15082-95. DOI https://doi.org/10.1039/D1NJ00925G
- Islam, S.M.; Havranek, B.; Ibnat, Z.; and Roy, P.N. “New insights into the role of hydrogen bonding in furanoside binding to protein” J. Phys. Chem. B, 2020, 124, 1919
- Qi, Y.; Lee, J.; Cheng, X.; Shen, R.; Islam, S.M.; Roux, B. and Im, W. “CHARMM-GUI DEER facilitator for spin-pair sistance distribution calculations and preparation of reMD simulations.” J. Comput. Chem., 2020, 1-9. https://doi.org/10.1021/acs.jpcb.9b11924
- Cheng, L.; Islam, S.M.; Mankad, N., “Synthesis of allylic alcohols via Cu-catalyzed hydrocarbonylative coupling of alkynes with alkyl halides” J. Amer. Chem. Soc. 2018, 140 (3), 1159–1164. https://doi.org/10.1021/acs.joc.2c02746
- Matthies, D.; Dalmas, O; Borgnia, M.J.; Dominik, P.; Merk, A.; Rao, P.; Reddy, B.; Islam, S.M.; Bartesaghi, A.; Perozo, E. and Subramaniam, S. “Cryo-EM Structures of the Magnesium Channel CorA Reveal Symmetry Break upon Gating” Cell, 2016, 164(4), 747-756. https://doi.org/10.1016/j.cell.2015.12.055
- Islam, S.M. and Roux, B. “Simulating the distance distribution between spin-labels attached to proteins” J. Phys. Chem. B, 2015, 119(10), 3901-3911. https://doi.org/10.1021/jp510745d
- Kazmier, K.; Sharma, S.; Islam, S.M.; Roux, B.; Mchaourab, H.S. “Conformational cycle and ion-coupling mechanism of the Na+/hydantoin transporter Mhp1.” PNAS, 2014, 111, 14752-14757. https://doi.org/10.1073/pnas.1410431111
- Jo, Sunwan; Cheng, X.; Islam, S.M.; Huan, L.; Han, W.; Roux, B.; MacKerell, A.D.; Im, W. “Chapter eight CHARMM-GUI PDB manipulator for advanced biomolecular modeling and simulations” Advances in Protein Chemistry and Structural Biology, 2014, 96, 235-264. https://doi.org/10.1016/bs.apcsb.2014.06.002
- Kazmier, K.; Sharma, S.; Quick, M.; Islam, S.M.; Weinstein, H.; Javitch, J.A.; Roux, B.; Mchaourab, H.S. “Conformational dynamics of ligand-dependent alternating access in LeuT” Nature Structural & Molecular Biology, 2014, 21, 472-479. https://doi.org/10.1038/nsmb.2816
- Raghuraman, H.; Islam, S.M.; Mukhergie S.; Roux, B. and Perozo, E. “Dynamic transitions at the outer vestibule of the KcsA potassium channel during gating”, PNAS, 2014, 111, 1831-1836. https://doi.org/10.1073/pnas.1314875111
- Islam, S.M.; Stein, A.R.; Mchaourab, H. and Roux, B. “Structural refinement from Restrained-Ensemble simulations based on EPR/DEER data: application to T4 Lysozyme” J. Phys. Chem. B, 2013, 117, 4740-4754. https://doi.org/10.1021/jp311723a
- Islam, S.M.; Richards, M.; Taha, H.; Lowary, T.L. and Roy, P.N. “Conformational analysis of oligo-arabinofuranosides: overcoming torsional barriers with umbrella sampling” J. Chem. Theory Comp., 2011, 7(9), 2989–3000. https://doi.org/10.1021/ct200333p
PROFESSIONAL ACTIVITIES AND SCHOLARLY SERVICES
Reviewer
- Journal of Physical Chemistry
- Journal of Chemical Information and Modeling
- Communications Biology
- Nature Protocols
- Microbial Risk Analysis
- ACS Omega
- Structure
- Biology
- Molecules
Professional Society Memberships
- Biophysical Society
- American Chemical Society
Grants
- Agency: NIH, Grant number: P30 GM145765, Role on grant: Pilot PI, Dates: 09/2023-08/2025, Amount: $100,000, Title: Finding and evaluating mutations in phenylalanine-4-hydroxylase (PAH) to develop targeted therapeutics for Phenylketonuria (PKU)
- Agency: NIH, Grant number: U54 MD015959, Role on grant: Research Core PI, Dates: 09/2023-08/2027, Amount: $1,600,583, Title: Integrating Low-Cost Paper-Based Devices and Personalized Immunotherapeutics to Treat Triple Negative Breast Cancer
- Agency: NIH, Grant number: 3U54MD015959-02S1, Role on grant: Co-PD, Dates: 09/2023-08/2025, Amount: $598,891, Title: Interdisciplinary Health Equity Research (IHER) Center - DSU Data Science Research and Education Hub
- Agency: NSF ACCESS, Grant number: 2138259/CHE210078, Role on grant: PI, Dates: 09/2022-01/2025, Amount: National supercomputer access equivalent to $1,45,000, Title: Computational design and optimization of small molecule and protein inhibitors for the use against variants of SARS-CoV-2
RECENT COURSES TAUGHT
- Biochemistry
- General Chemistry
- Physical Chemistry
- Computational (Bio)Chemistry